Dear Sen4Cap and @cudroiu ,
I have changed configuration of L4B accordingly and seems step 1 and 2 are working. It’s stuck at step 3 and I got below errors. Please help to solve it out.
Thanks and Regards,
Henry
Traceback (most recent call last):
File “/usr/share/sen2agri/S4C_L4B_GrasslandMowing/Bin/src_s2/S2_main.py”, line 971, in
main()
File “/usr/share/sen2agri/S4C_L4B_GrasslandMowing/Bin/src_s2/S2_main.py”, line 965, in main
do_cmpl=s4cConfig.do_cmpl, test=s4cConfig.test)
File “/usr/share/sen2agri/S4C_L4B_GrasslandMowing/Bin/src_s2/S2_main.py”, line 740, in main_run
test=test)
File “/usr/share/sen2agri/S4C_L4B_GrasslandMowing/Bin/src_s2/S2_main.py”, line 222, in run_proc
burned_pixels, seg_attributes = S2_gmd.layer2mask(segmentsFile, output_vrt, segmentsOutRaster, layer_type=‘segments’, options=options_layer_burning)
File “/usr/share/sen2agri/S4C_L4B_GrasslandMowing/Bin/src_s2/S2_gmd.py”, line 416, in layer2mask
ogr_data.ExecuteSQL(‘CREATE SPATIAL INDEX ON %s’ % LayerName)
File “/home/sen2agri-service/miniconda3/envs/sen4cap/lib/python3.6/site-packages/osgeo/ogr.py”, line 800, in ExecuteSQL
return _ogr.DataSource_ExecuteSQL(self, *args, **kwargs)
RuntimeError: CreateSpatialIndex : unsupported operation on a read-only datasource.
Hi,
I received the exact same error. Did you find a way to solve this problem?
Best regards
Bastian
No, Bastian. I am still waiting for sen4cap support. Hope it can be solved out.
Cheers!
@kraftek do you think there will be a solution to this problem soon? We were planning on using the mowing results next week…
Dear Bastian,
I assume that you are running on a local machine and you cannot provide me access to it in order to have a look over it. In this case:
- can you provide the config file you used?
- In a console, could you login under the user sen2agri-service (sudo su -l sen2agri-service) and execute the commands:
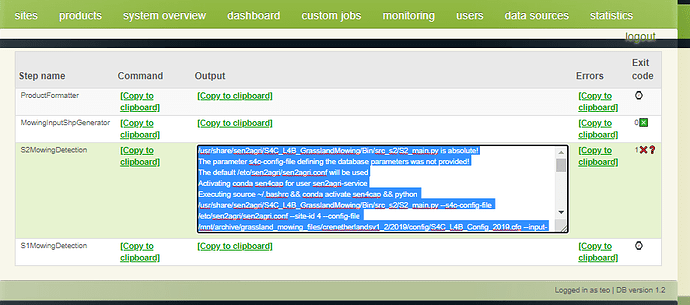
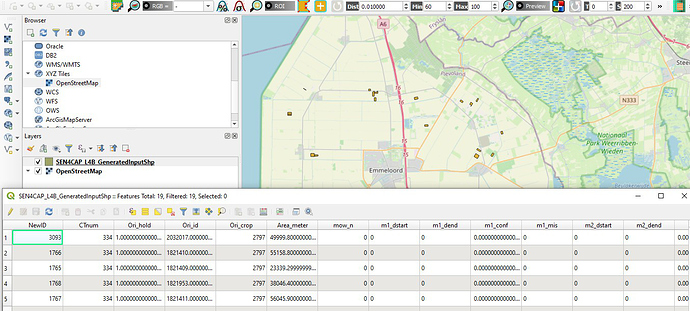
- Corresponding to the MowingInputShpGenerator step - check that file corresponding to the -o parameter is correctly created and can be opened in QGis
- If 1. was OK, execute the command corresponding to the S2MowingDetection step.
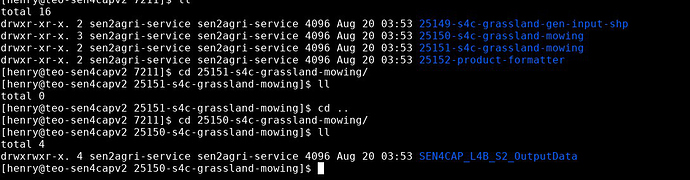
(Before executing the above steps commands please ensure that the paths inside the commands like /mnt/archive/orchestrator_temp/s4c_l4b/XXXXXX/XXXXXX-s4c-grassland-gen-input-shp, /mnt/archive/orchestrator_temp/s4c_l4b/XXXXX/XXXXX-s4c-grassland-mowing/SEN4CAP_L4B_S2_OutputData and /mnt/archive/orchestrator_temp/s4c_l4b/XXXXX/XXXXX-product-formatter/ are created - you can create them using mkdir -p < PATH > ).
Please provide the command for this step and the logs that resulted during execution.
Best regards,
Cosmin
Dear @cudroiu,
Welcome back.
We are running Netherlands sample site and I have attached for L4B Config.
Step 1 is working fine and I can view the shape file in QGIS. I checked all those folders and they are there. Refer to below screenshot. I also attached the command and result log. Let me know if you need further info to solve it out.
S4C_L4B_NLD_Config.zip (1.8 KB)
l4b_commandANDlog.txt (31.7 KB)
Thanks and Regards,
Henry
Hello,
It seems that some updates were made in the gdal version from the conda (I assume there is a newly installed or updated machine) and in this new version were introduced some constraints.
I am attaching 2 patches (S2_gmd.patch and S1_gmd.patch in the archive Sen4CAP_v_1_2_L4B_S1_gmd_patches.zip (790 Bytes) ) for this issue that can be installed with the following commands:
unzip Sen4CAP_v_1_2_L4B_S1_gmd_patches.zip
sudo patch -u /usr/share/sen2agri/S4C_L4B_GrasslandMowing/Bin/src_s2/S2_gmd.py -i S2_gmd.patch
sudo patch -u /usr/share/sen2agri/S4C_L4B_GrasslandMowing/Bin/src_s1/S1_gmd.py -i S1_gmd.patch
Hope this helps.
Best regards,
Cosmin
2 Likes
Dear Cosmin,
It’s worked like a magic. Thanks a lot for your support as usual.
Regards,
Henry